Generative Adversarial Networks for Biological Image Synthesis
Abstract
Generative Modelling, a branch of Machine Learning that focuses on generating realistic-looking samples, has traditionally constituted the upper bound of what Machine and Deep Learning models can achieve. This regime has completely changed in recent years, especially after 2014, when I. Goodfellow presented a generative model comprising two competing neural networks: Generative Adversarial Network (GAN) [8]. Subsequently, a plethora of models based on GAN have been proposed with impressive results.
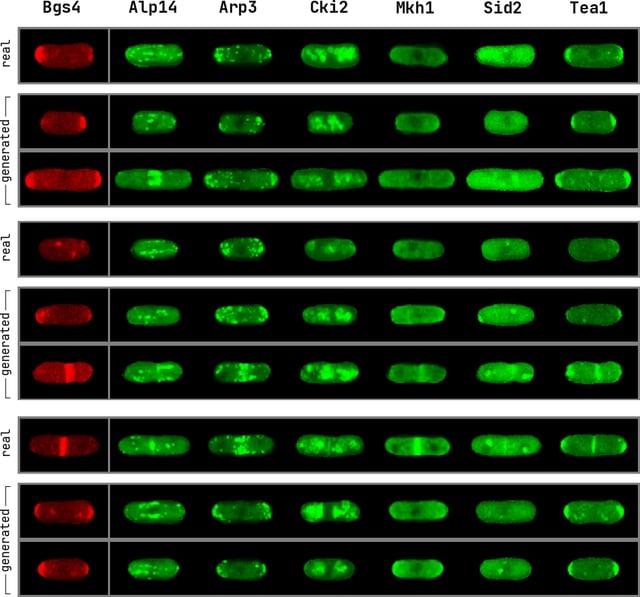
Concurrently, more and more research is devoted to developing techniques for demystifying biological functions at a cellular level. Among its purposes is creating artificial intelligence systems that provide insights into how different proteins operate and the way their co-location is connected with higher-level functions as captured in spectroscopic techniques. In an endeavor to apply modern machine learning techniques to synthesize cells imaged with fluorescent microscopy, in this project we employ Generative Adversarial Networks.
In particular, we use the network architectures presented by Osokin et al. [19] to generate realistic images of yeast fission cells imaged by fluorescent microscopy, aspiring that by being able to do so, the networks must capture the correlations present in the localization of the various proteins of interest. In addition, these models are able to generate multichannel images and therefore circumvent one major limitation of fluorescent microscopy.
Keywords
GAN, Fluorescent Microscopy, DCGAN, Wasserstein Loss, Gradient Penalty, FID, IS, Classifier 2-Sample Test, Yeast Fission, Cell Image Synthesis, GFP, RFP